

| RQV30029 | |
| I. Background | |
hCov-NL63 was first isolated from an aspirate from a 7-month-old baby in the Netherlands in early 2004. Since then, HCoV-NL63 infection has been proven to be a universal disease worldwide and is related to many clinical symptoms. Diagnosis includes severe lower respiratory tract infection, buttitis and bronchiolitis. HCoV-NL63 causes childhood diseases. The elderly and immunocompromised children account for 1.0-9.3% of childhood respiratory infections. 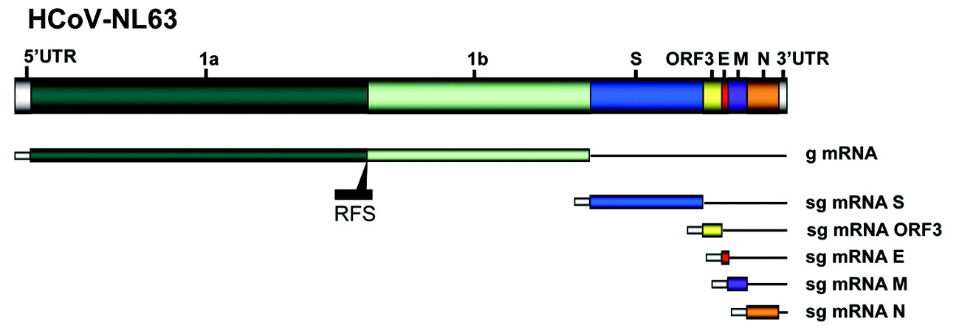
According to the introduction of CDC and WHO, nucleic acid diagnosis for HCoV-NL63 is generally for RdRP (RNA-dependent RNA polymerase), Gene N and Gene E, and Gene S is more used for the development of corresponding drugs and vaccines. The NCBI included sequence is JX504050.1, with a total length of 27553bp, and the gene encodes 4 structural proteins: S, M, N and E. In the past, the performance evaluation of the detection limit of the kit generally used in vitro transcribed RNA or clinically positive virus as the standard product for performance evaluation. However, both have obvious certainty. First, the RNA transcribed in vitro is generally very pure and single, does not involve extraction, and does not meet the microenvironment of the clinical virus. Therefore, the detection sensitivity will inevitably be overestimated. This is also a high probability of false The main reason for the negative; clinically positive virus again, even if it is an inactivated virus, still has a great biological safety hazard. Therefore, the laboratory level is required to be P3-P4. Most of the units that develop diagnostic kits do not meet the requirements. The source of clinically positive viruses is also very limited and cannot be stably supplied, and is repeatedly used for performance evaluation of the kit. | |
| II.product description | |
It has the complete envelope structure of the virus and the nucleic acid sequence corresponding to HCoV-NL63, but the pseudovirus standard product with low biosafety risk perfectly overcomes the shortcomings of choosing in vitro transcribed RNA or clinically positive virus as the standard product. It is very suitable for the standard product of nucleic acid diagnostic performance evaluation. Construction process: HCoV-NL63 gene synthesis → pseudovirus packaging → purification → QC detection (ddPCR) | |
| III. product description | |
| product name: | hCov-NL63-ORF1ab/RdRP Pseudovirus Standard Reference |
| Corresponding sequence (see appendix for details): | ORF1ab/RdRP(2780bp)、Gene N(1134bp)、Gene E(234bp) |
| Specifications: | 1ml |
| Number of copies: | |
| Biosafety level: | P2 |
| Transportation and storage methods: | Dry ice transportation, -80°C save |
| Validity period: | |
| IV. Product usage and advantages | |
| Product Usage: | HCoV-NL63Performance evaluation of diagnostic kits |
| Product advantages: | ·The pseudovirus standard of HCoV-NL63 is different from RNA synthesized in vitro and clinically positive live virus. It perfectly overcomes the shortcomings of both, and is really suitable for performance evaluation of the kit, including detection limit, specificity, repeatability, etc. ; ·According to the characteristics of the HCoV-NL63 coronavirus, a total of 3 segments of ORF1ab/RdRP (RNA-dependent RNA polymerase), Gene E (envelope protein), and Gene N (nucleocapsid phosphoprotein) were selected for the packaging of pseudovirus , 3 segments well represent the characteristic sequence of HCoV-NL63; ·Coby Bio has designed the primers and probe sequences of the ddPCR system, which can complete the QC copy number detection of pseudovirus without constructing a standard curve, which can very accurately characterize the copy number of the standard product, and overcome the Q-PCR analysis of CT Inaccuracy of the relative judgment standard of the value; ·Pseudovirus standard products, non-pathogenic, renewable, reliable quality control methods, stable between batches, can be prepared and supplied stably for a long time, and require laboratory biosafety level P2 to meet the safety needs of many organizations. |
| Ⅴ. appendix | |
ORF1ab/RdRP: 1 agtgttgaca tttcttattt aaacgagcaa ggggttctag tgcagctcga ctagaaccct 61 gtaatggcac ggacatcgat aagtgtgttc gtgcttttga catttataat aaaaatgttt 121 cattcttggg taagtgtttg aagatgaact gtgttcgttt taaaaatgct gatcttaagg 181 atggttattt tgttataaag aggtgtacta agtcggttat ggaacacgag caatccatgt 241 ataacctact taacttttct ggtgctttgg ctgagcatga tttctttact tggaaagatg 301 gcagagtcat ttatggtaat gttagtagac ataatcttac taaatatact atgatggact 361 tggtctatgc tatgcgtaac tttgatgaac aaaattgtga tgttctaaaa gaagtattag 421 ttttaactgg ttgttgtgac aattcttatt ttgatagtaa gggttggtat gacccagttg 481 aaaatgaaga tatacataga gtttatgcat ctcttggcaa aattgtagct agagctatgc 541 ttaaatgcgt tgctctatgt gatgcgatgg ttgctaaagg tgttgttggt gttttaacat 601 tagataacca agatcttaat ggtaactttt atgattttgg tgattttgtt gttagcttac 661 ctaatatggg tgttccctgt tgtacatcat attattctta tatgatgcct attatgggtt 721 taactaattg tttagctagt gagtgttttg tcaagagtga tatttttggt agtgatttta 781 aaacttttga tttgcttaag tatgatttca ctgaacataa agaaaattta ttcaataagt 841 actttaagca ttggagtttt gattatcatc ctaattgtag tgactgttat gatgatatgt 901 gtgttataca ttgtgctaat tttaatacac tatttgctac aactatacca ggtactgctt 961 ttggtccact atgtcgtaaa gtttttatag atggtgttcc acttgttaca actgctggtt 1021 atcattttaa gcaattaggt ttggtttgga ataaagatgt taacacacac tcagttaggt 1081 tgacaattac tgaacttttg caatttgtca ccgacccttc cttgataata gcttcttccc 1141 cagcactcgt tgatcaacgc actatttgtt tttctgttgc agcattgagt actggtttga 1201 caaatcaagt tgttaagcca ggtcatttta atgaagagtt ttataacttt cttcgtttaa 1261 gaggtttctt tgatgaaggt tctgaactta cattaaaaca tttcttcttc gcacagaatg 1321 gtgatgctgc tgttaaagat tttgactttt accgttataa taagcctacc attttagata 1381 tttgtcaagc tagagttaca tataagatag tctctcgtta ttttgacatt tatgaaggtg 1441 gctgtattaa ggcatgtgaa gttgttgtaa caaatcttaa taagagtgct ggttggccat 1501 taaataagtt tggtaaagct agtttgtatt atgaatctat atcttatgaa gaacaggatg 1561 ctttgtttgc tttgacaaag cgtaatgtcc tccctactat gacacagctg aatcttaagt 1621 atgctattag tggtaaagaa cgtgctagaa ctgttggtgg tgtttctctg ttgtctacaa 1681 tgaccacaag acaataccat caaaaacatc ttaaatccat tgttaataca cgcaatgcca 1741 ctgttgttat tggtactacc aaattttatg gtggttggaa taatatgttg cgtactttaa 1801 ttgatggtgt tgaaaaccct atgctcatgg gttgggatta tcccaaatgt gatagagctt 1861 tgcctaacat gatacgtatg atttcagcca tggtgttggg ctctaagcat gttaattgtt 1921 gtactgcaac agataggttt tataggcttg gtaatgagtt ggcacaagtt ttaacagaag 1981 ttgtttattc taatggtggt ttttatttta agccaggtgg tacgacttct ggtgacgcta 2041 gtacagctta tgctaattct atttttaaca tttttcaagc cgtgagttct aacattaaca 2101 ggttgcttag tgtcccatca gattcatgta ataatgttaa tgttagggat ctacaacgac 2161 gtctgtatga taattgctat aggttaacta gtgttgaaga gtcattcatt gatgattatt 2221 atggttatct taggaaacat ttttcaatga tgattctctc tgatgacggt gttgtctgtt 2281 ataacaagga ttatgctgag ttaggttata tagcagacat tagtgctttt aaagccactt 2341 tgtattacca gaataatgtc tttatgagta cttctaaatg ttgggttgaa gaagatttaa 2401 ctaagggacc acatgagttt tgttcccagc atactatgca aatagttgac aaagatggta 2461 cctattattt gccttaccca gatcctagta ggatcttgtc agctggtgtt tttgttgatg 2521 atgttgttaa gacagatgct gttgttttgt tagaacgtta tgtgtcttta gctattgatg 2581 cataccctct ttcaaaacac cctaattctg aatatcgcaa ggttttttac gtattacttg 2641 attgggttaa gcatcttaac aaaaatttga atgagggtgt tcttgaatct ttttctgtta 2701 cacttcttga taatcaagaa gataagtttt ggtgtgaaga tttttatgct agtatgtatg 2761 aaaattctac aatattgcaa
Gene N: ATGGCTAGTGTAAATTGGGCCGATGACAGAGCTGCTAGGAAGAAATTTCCTCCTCCTTCATTTTACATGCCTCTTTTGGTTAGTTCTGATAAGGCACCATATAGGGTCATTCCCAGGAATCTTGTCC CTATTGGTAAGGGTAATAAAGATGAGCAGATTGGTTATTGGAATGTTCAAGAGCGTTGGCGTATGCGCAGGGGGCAACGTGTTGATTTGCCTCCTAAAGTTCATTTTTATTACCTAGGTACTGGACC TCATAAGGACCTTAAATTCAGACAACGTTCTGATGGTGTTGTTTGGGTTGCTAAGGAAGGTGCTAAAACTGTTAATACCAGTCTTGGTAATCGCAAACGTAATCAGAAACCTTTGGAACCAAAGTTC TCTATTGCTTTGCCTCCAGAGCTCTCTGTTGTTGAGTTTGAGGATCGCTCTAATAACTCATCTCGTGCTAGCAGTCGTTCTTCAACTCGTAACAACTCACGAGACTCTTCTCGTAGCACTTCAAGAC AACAGTCTCGCACTCGTTCTGATTCTAACCAGTCTTCTTCAGATCTTGTTGCTGCTGTTACTTTGGCTTTAAAGAACTTAGGTTTTGATAACCAGTCGAAGTCACCTAGTTCTTCTGGTACTTCCAC TCCTAAGAAACCTAATAAGCCTCTTTCTCAACCCAGGGCTGATAAGCCTTCTCAGTTGAAGAAACCTCGTTGGAAGCGTGTTCCTACCAGAGAGGAAAATGTTATTCAGTGCTTTGGTCCTCGTGAT TTTAATCACAATATGGGGGATTCAGATCTTGTTCAGAATGGTGTTGATGCCAAAGGTTTTCCACAGCTTGCTGAATTGATTCCTAATCAGGCTGCGTTATTCTTTGATAGTGAGGTTAGCACTGATG AAGTGGGTGATAATGTTCAGATTACCTACACCTACAAAATGCTTGTAGCTAAGGATAATAAGAACCTTCCTAAGTTCATTGAGCAGATTAGTGCTTTTACTAAACCCAGTTCTATCAAAGAAATGCA GTCACAATCATCTCATGTTGCTCAGAACACAGTACTTAATGCTTCTATTCCAGAATCTAAACCATTGGCTGATGATGATTCAGCCATTATAGAAATTGTCAACGAGGTTTTGCATTAA Gene E: ATGTTCCTTCGATTAATTGATGACAATGGTATTGTCCTCAATTCCATTTTATGGCTCCTTGTTATGATATTTTTCTTTGTGTTGGCAATGACCTTTATTAAACTGATTCAATTGTGTTTTACTTGTC ATTATTTTTTTAGTAGGACATTATATCAACCAGTTTATAAAATTTTTCTTGCTTACCAAGATTATATGCAAATAGCACCTGTTCCAGCTGAAGTACTAAATGTCTAA | |