[Standard product] RNA-Fusion Cocktail is upgraded again, FFPE slide free trial
Gene fusion refers to linking the coding regions of two or more genes end to end and placing them under the control of the same set of regulatory sequences (including promoters, enhancers, and terminator, etc.) to form a chimeric gene. Gene fusions are usually caused by chromosomal rearrangements. Abnormal gene fusion events can cause malignant hematological diseases and tumors. Therefore, analyzing gene fusion phenomena will help to explore the pathogenesis and biomaker screening, which is of great clinical significance.
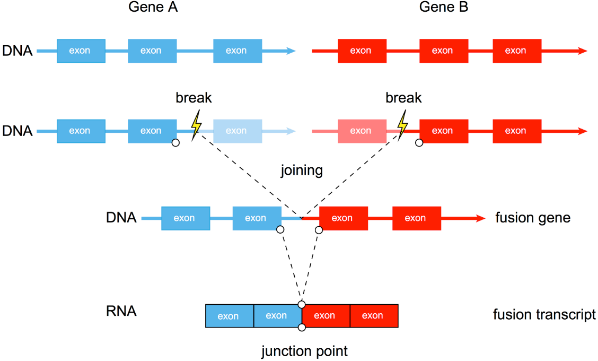
There are three main mechanisms for gene fusion, as shown in the figure below:
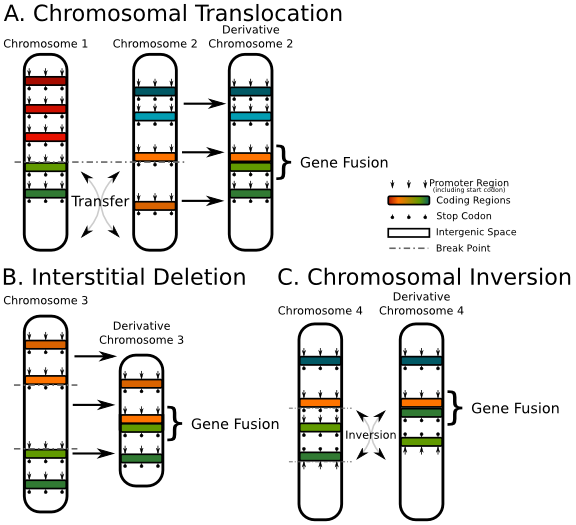
Fig 2. Three common mechanisms of gene fusion
Three common mechanisms of gene fusion:
1) Chromosomal Translocation, chromosomal translocation. As shown in Figure A above, the two fragments on chromosomes 1 and 2 are cross-exchanged, resulting in the fusion of the light green gene on chromosome 1 and the orange gene on chromosome 2;
2) Interstitial deletion. As shown in the figure above, the segment between the orange gene and the light green gene on chromosome 3 is deleted, which eventually leads to the fusion of these two genes;
3) Chromosomal Inversion, chromosome inversion. For example, the fragment between the orange gene and the dark green gene on chromosome 4 is inverted, which eventually leads to the fusion of the orange gene and the light green gene.
The relationship between gene fusion and cancer
Past studies have consistently shown that gene fusion is closely related to the occurrence and development of various diseases, especially cancer, and even the direct cause of some cancers. Therefore, gene fusion has also become an important research content in the current omics big data analysis.
At present, it has been reported that many cancers are closely related to gene fusion, as shown in the following table:
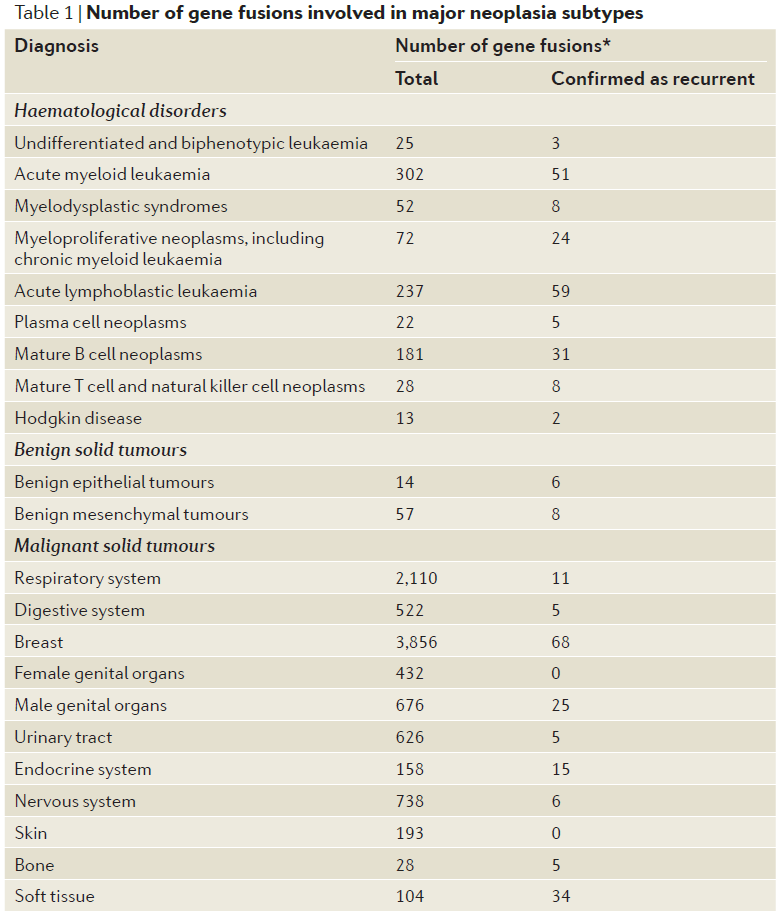
Table 1. The relationship between tumors and fusion genes
Identification of gene fusions based on whole-genome sequencing and transcriptome sequencing
The identification of gene fusion can be based on whole-genome sequencing (WGS), transcriptome sequencing data (RNA-seq), or a better combination of the two technologies.
The gene fusion identified by whole-genome sequencing can basically be determined to be caused by a certain mutation at the genome level, but if there is no transcriptome sequencing data, it is impossible to accurately determine whether the new gene generated after the fusion can be expressed, or the amount of expression. High and low.
The gene fusion identified by transcriptome sequencing data can be clearly expressed as a gene fusion, but it cannot be completely determined whether it is caused by genomic mutation or the RNA fusion that occurs after the transcription of two different genes.
Therefore, if conditions permit, combining whole genome sequencing (WGS, or panel) and transcriptome sequencing (RNA-seq) to identify gene fusions can obtain more accurate identification results.
RNA-Fusion Panel is upgraded again
Kebai Biosciences launched the 11-site RNA-Fusion Cocktail control in 2020, which is widely used in customer quality control testing. This product contains 11 common fusion sites and is a mixed-sample panel of RNA, all of which are calibrated by ddPCR .
In 2021, based on the demands of our customers, we upgraded our RNA-Fusion Cocktail control, from 11 sites to 22 sites, and quality control products for more sites, the format was upgraded from RNA solution to FFPE slide, and more Simulating clinical samples, involving extraction and other processes, all upgraded products use ddPCR to calibrate the copy number.
↓↓↓↓↓↓
Panel-Ref® RNA-Fusion Cocktail FFPE Control RQP90036
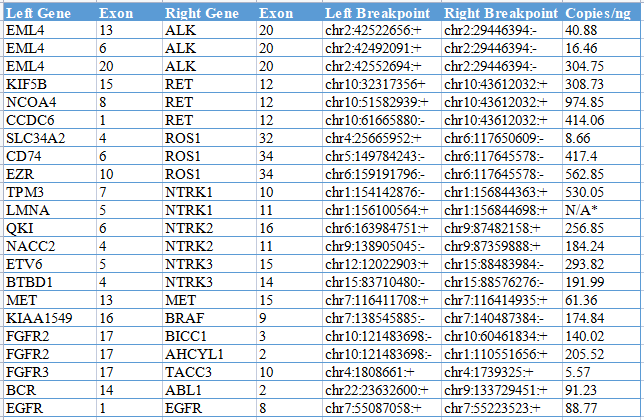
*LMNA-NTRK1 is being tested by ddPCR

